Cancer
Cancer has been established as the most common cause of mortality in Australia. It is estimated that 115,000 people will be diagnosed with the disease in 2010, and more than 43,000 are expected to die from cancer (Cancer Council Australia; http://www.cancer.org.au/aboutcancer/FactsFigures.htm). Changes to cellular DNA cause the emergence of cancer cells in the body, which are defined by their uncontrolled growth, and their ability to invade and spread throughout the body. Normal cells in the body follow a defined cycle of growth, division, and programmed cell death (apoptosis). When this process breaks down, cancer can form. Unlike normal cells, cancer cells do not experience programmatic death and instead continue to grow and divide, resulting in a mass of abnormal cells growing out of control.
Current treatments are capable of masking symptoms and delaying death, sometimes providing temporary remission. However, more effective targeted therapies are required to mount a more efficient defence against this disease, and to combat “resistance” to current therapies. Our objective is to provide model cellular systems which allow us to increase our knowledge and understanding of the tumour micro-environment and disease progression, as well as providing platforms to improve clinical predictability of lead compounds during early drug discovery.
The current areas of cancer research in Discovery Biology include Prostate, Breast and Pancreatic cancer. A variety of in vitro methodologies including the utilisation of 3D culturing techniques and high content imaging systems are being used to investigate the mechanisms of cellular interactions, cancer growth, progression and prevention, and the identification of compounds which specifically act upon these.
Drug development is a risky, time consuming and expensive exercise, therefore, it is imperative to select the right compound early in the process. A platform that allows selection of biologically relevant compounds with a high probability of converting their in vitro activity into in vivo activity, would be highly beneficial to the drug discovery process, providing decreased attrition rates and time to the clinic. Such a model would be robust, automated, mimic the complexity of the biological system with multiple pathways and measure disease-relevant cellular responses.
High Content Screening (HCS) allows increased understanding of the effects of compounds on the target and of the associated cell biology and predictive toxicology. 3D cell culture mimics the in vivo environment more realistically than 2D monolayers, allowing for the subtle interplay of cells of the same or different origins within a support network. Combining the two we are developing a platform to evaluate multiple cancers from a variety of levels.
Small Molecule Drug Discovery
Cancer Therapeutics CRC (CTx) (https://www.cancercrc.com) worked closely with leading Australian and international researchers and institutes to discover and develop small molecule discoveries into pre-clinical drug candidates for the next generation of global cancer therapies. Discovery Biology played an integral part of CTx, with expertise in molecular and cellular biology, drug discovery, and pharmacology.
Discovery Biology develops and undertakes high throughput screening (HTS) assays for the identification of active compounds from large chemical libraries, against candidate cancer targets. Following hit identification, we also perform lead generation and optimization by means of additional biochemical and cellular assays.
Breast Cancer
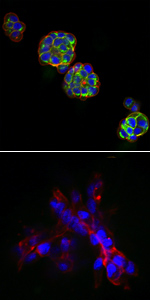 In vitro 3D Modelling of Breast Cance
In vitro 3D Modelling of Breast Cance
Breast cancer is a heterogeneous disease where treatment is dependent on tumour characteristics. Characteristics of tumours such as size, grade, metastases and receptor overexpression status are important in defining therapy for breast cancer patients. Breast cancer patients may be treated with one or more chemotherapeutics and/or targeted therapy. However, patient’s responses to breast cancer therapies are variable. Thus, further characterisation of the tumour microenvironment and drug effects are required.
3D cell culture models for breast cancer have been developed to create a more in vivo-like environment to facilitate further research into areas of cell-cell and cell-extracellular matrix interactions in the presence of breast cancer therapeutics. The 3D cell culture models were developed in a miniaturised format (384- and 1536-well microtitre plate) and have been characterised for use in the screening of drug candidates and combination therapy. Furthermore, various drug resistance mechanisms have been identified utilising 3D breast cancer cell culture models. Further evaluation of potential therapeutics identified and chemotherapeutic drug resistance mechanisms are ongoing.
Pancreatic Cancer
In vitro 3D Modelling of Pancreatic Cancer
Pancreatic cancer is a disease with a very poor prognosis and today remains one of the most lethal human cancers. There is no way of early detection and few effective treatment options. Recent advances in treatments have only increased survival times by months, with optimal palliation being the goal. There is currently an urgent need for developing a detailed understanding of the molecular biology of the cancer. The research in Discovery Biology will help establish tools that may be used to prioritise strategies for the development of novel treatments.
This project aims to develop an innovative biological tumour model to study pancreatic cancer in a way that more closely mimics its in vivo environment. A 3D culture model will be used to gain new insights into normal and tumour pancreatic biology and facilitate further research into areas of cellular interactions and drug discovery. 3D culture models will be developed using commercial matrices, with a range of pancreatic cancer cell lines and compared with conventional 2D monolayer cultures. Markers examining cell proliferation, morphology and expression of various signalling pathways relevant to pancreatic cancer have been assessed, as have compounds known to have alternative anti-cancer properties.
Prostate Cancer
Regulation of CXCR4 by the Androgen Receptor in Prostate Cancer
The mortality in prostate cancer (PCa) is attributed to bone metastasis, and the ability of cells to migrate is predominantly mediated by chemokines and their receptors. Of these, CXC Chemokine Receptor 4 (CXCR4) has been shown to regulate the metastasis of PCa tumour cells, particularly to bone. The Androgen Receptor (AR) is known as a key regulator of PCa cell growth in the early stages of the disease, and has a role in cancer progression. Normal prostate and early-stage PCa cell lines depend on androgens and AR for  growth, and are termed Androgen Dependent (AD). Other cell lines are derived from Androgen Independent (AI) cancer populations, where they have evolved to grow independently of androgens and AR.
growth, and are termed Androgen Dependent (AD). Other cell lines are derived from Androgen Independent (AI) cancer populations, where they have evolved to grow independently of androgens and AR.
There is evidence for inter-regulation between these two receptors; over-expression of AR in the DU145 cell line results in down-regulation of CXCR4 mRNA expression, and an inhibition of cell migration to the CXCR4 ligand, Stromal-Derived Factor 1-alpha (SDF-1α). In this research project, we aim to further characterise the regulation of CXCR4 by AR in PCa cells, by investigating how different facets of CXCR4 function are affected by AR expression in AI and AD cell lines. Regulation of CXCR4 by AR has important implications for the development and progression of PCa. Further characterisation of the link between AR and CXCR4 expression and function is necessary to understand the intricate mechanisms through which PCa cells are able to metastasize to bone in the end stages of the disease.
Unravelling Prostate Cancer Pathways with High Content Imaging
Early stage, prostate-confined cancer is generally susceptible to androgen ablation by chemical or surgical treatment. Late stage, metastatic PCa however, is often fatal, as the tumour grows in an AI manner, rendering it more difficult to control. The pathways that lead to metastasis are not well understood and therefore there are no treatments to prevent tumour spread. This project aims to elucidate pathways that are potential targets for therapeutic intervention, particularly those leading to metastasis. High content imaging has been utilized to analyze both 2D and 3D prostate cell cultures for inhibition of cellular proliferation from various compound libraries and sources. Compounds observed to inhibit PCa growth have been assayed for their effects on known PCa-related pathways, as well as pathways selected as candidates, based on live cell tracking with fluorescent labels. Integral to this research is the establishment of 3D cellular models in a microtitre format and the associated image analysis capabilities. Algorithms to allow us to compare 2D with 3D environments are thus necessary.
Further to this, a variety of stable fluorescent cell lines have been created. These contain plasmid constructs of potential anti-PCa targets, tagged to fluorescent proteins. This allows for the rapid high content identification of novel compounds that directly and specifically inhibit the anti-PCa targets previously identified.